 Prof. Dr. Oliver Niehuis
Prof. Dr. Oliver Niehuis
Ecology, Evolutionary Biology, and Biodiversity
Albert Ludwig University
Institute of Biology (Zoology)
Hauptstraße 1
D-79104 Freiburg
Room: 1030 (office hours by prior arrangement)
Phone: ++49 / 761 / 203 - 2506
E-Mail: oliver.niehuis@biologie.uni-freiburg.de
Research Interests
|
Causes of and Constraints on Novel Trait Evolution
We apply a wide array of comparative approaches to shed light on the genomic, molecular, and morphological underpinnings of novel traits. These include ‘bouquets’ of chemical traits. Trait data form the basis for deriving hypotheses on the causes and constraints that shaped the evolution of novel traits. Traits of particular interest to us are those used by arthropods to perceive their environment (e.g., eyes, odorant receptors) or for intra- and interspecific communication (e.g., pheromones) in host-parasite relationships.
|
   |
Chemical Ecology
|
| Many arthropods depend on chemical cues and signals for their intra- and interspecific communication. We use gas chromatography and mass spectrometry in combination with behavioral experiments to shed light on the chemical composition and function of these traits. The resulting data form the basis for inferring the possible mechanisms and forces that could have triggered the evolution of these traits (e.g., genome duplication, chemical deception). |
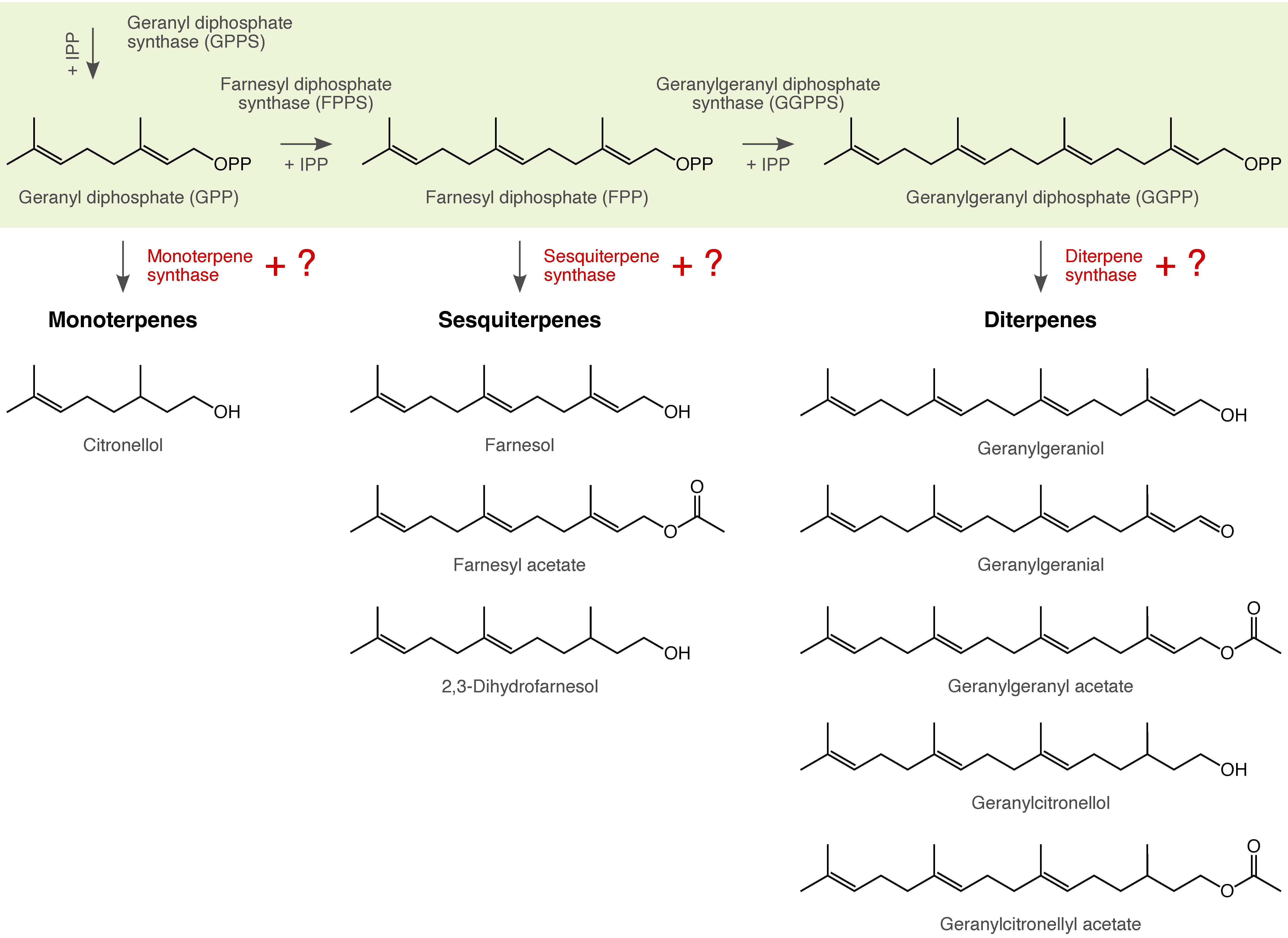
|
Morphology
|
| Modern 3-dimensional visualization techniques, such as micro-computed tomography, allow for high resolution visualization of morphological structures. We describe and compare morphological structures by qualitative and quantitative approaches. Their transformations in the course of evolution are reconstructed in selected taxa of arthropods. |
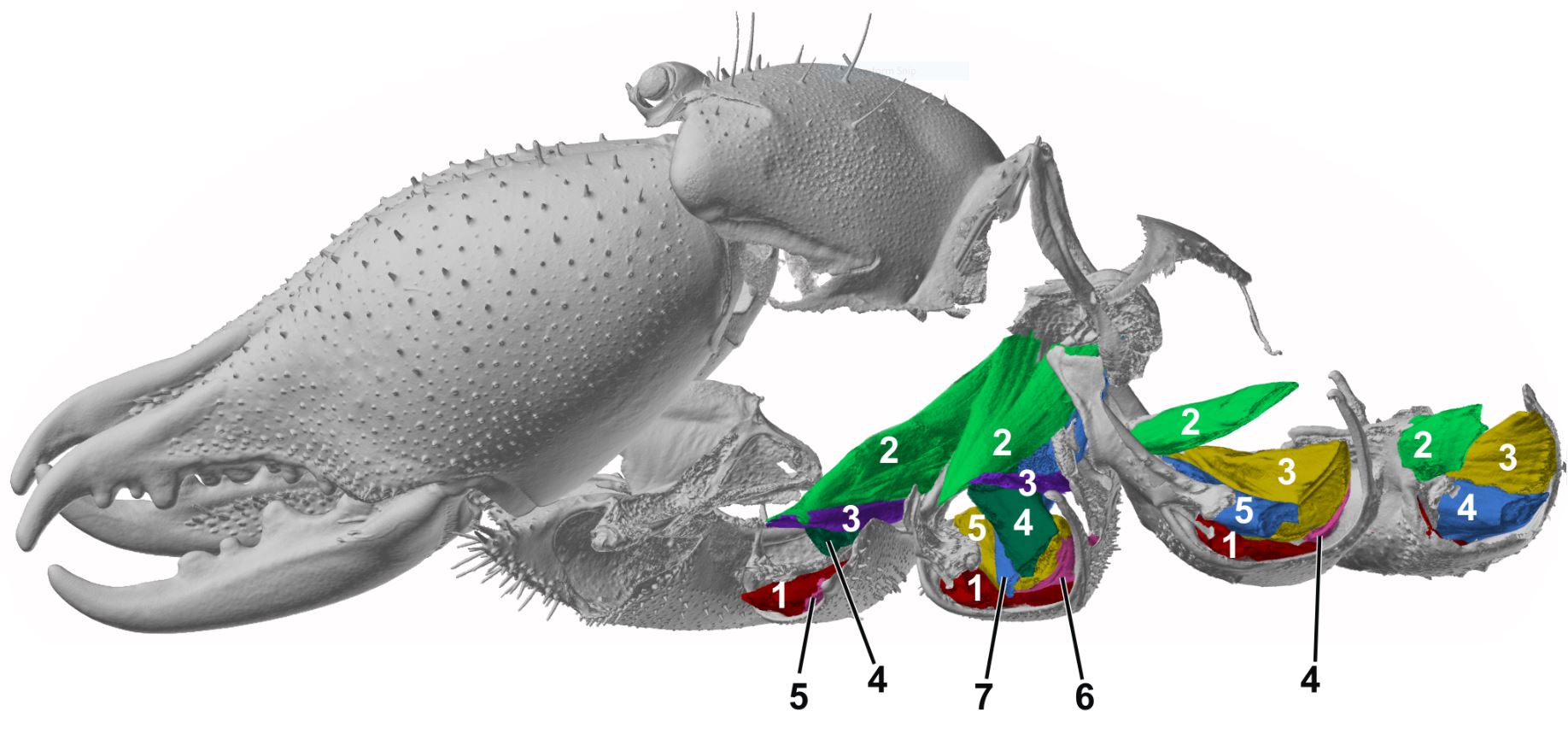 |
Comparative Genomics and Phylogenetics
|
| Comparing genomic data in a phylogenetic context is a powerful approach to identify genomic elements and mechanisms that could have triggered the evolution of novel traits. We apply this approach in combination with wet lab experiments (e.g., RNA interference) to elucidate the biosynthesis of novel chemical compounds and the evolution of new chemical profiles. |
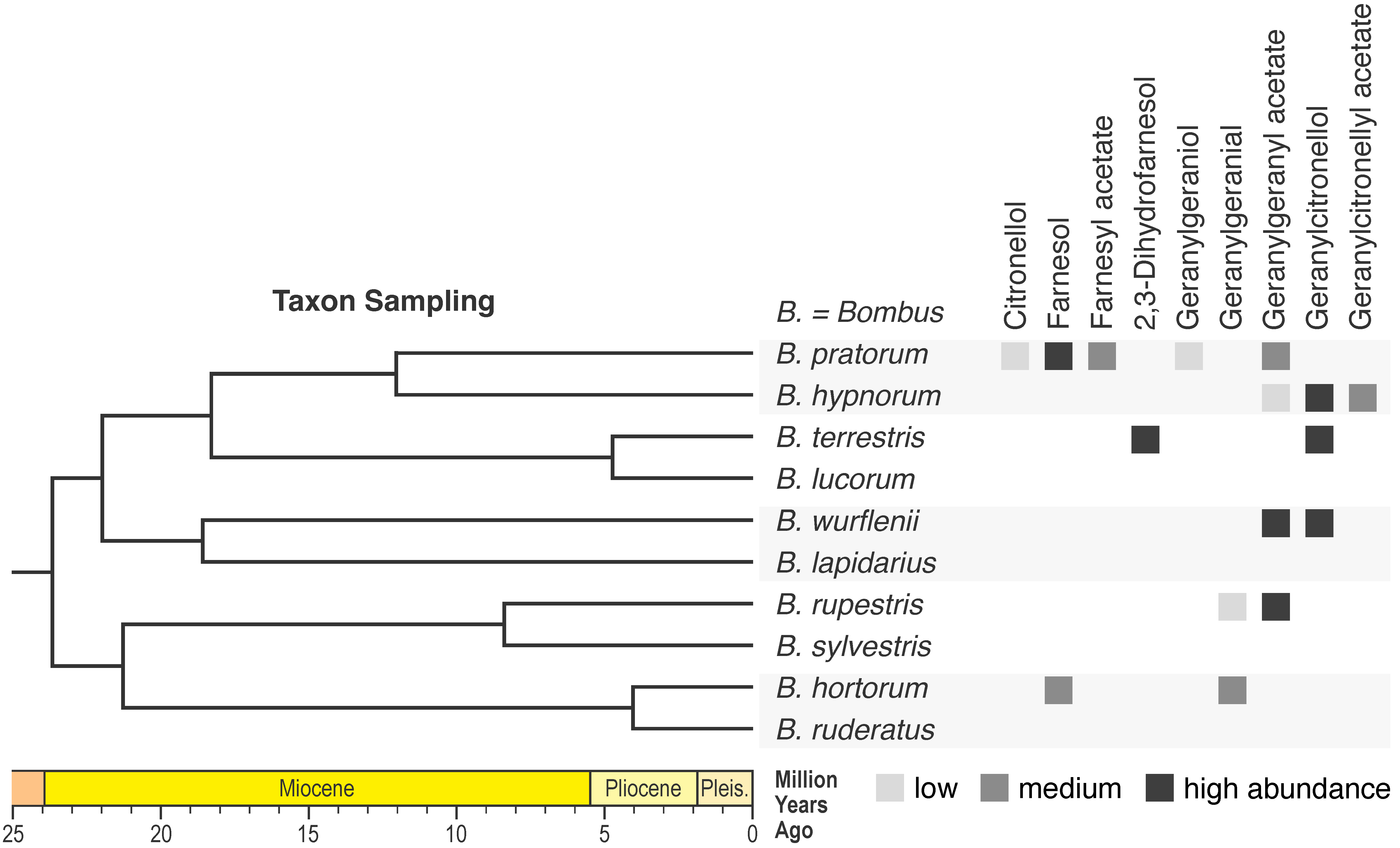 |
Bioinformatics
|
| Analyses of large datasets depend critically on bioinformatic tools. We participate in the development of open-source software for analyzing nucleotide sequence and mass spectrometric data. We rely in our daily work on the programming languages Julia and Python and annually offer a course (PM-24) to Bachelor students in which we teach the use of Python for solving bioinformatic tasks. |
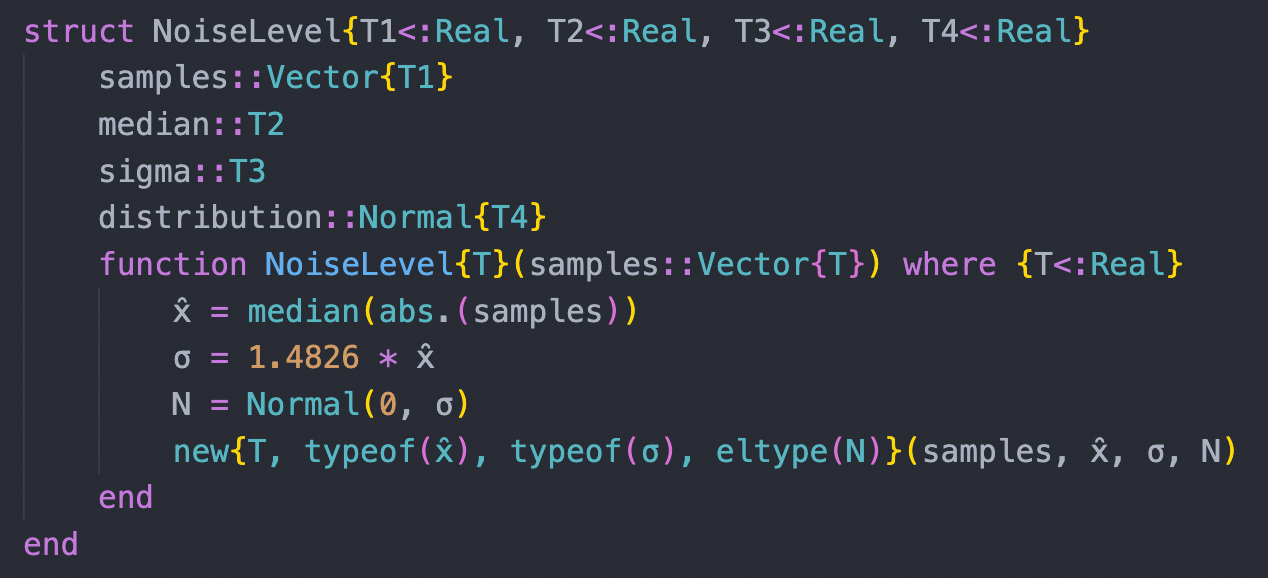 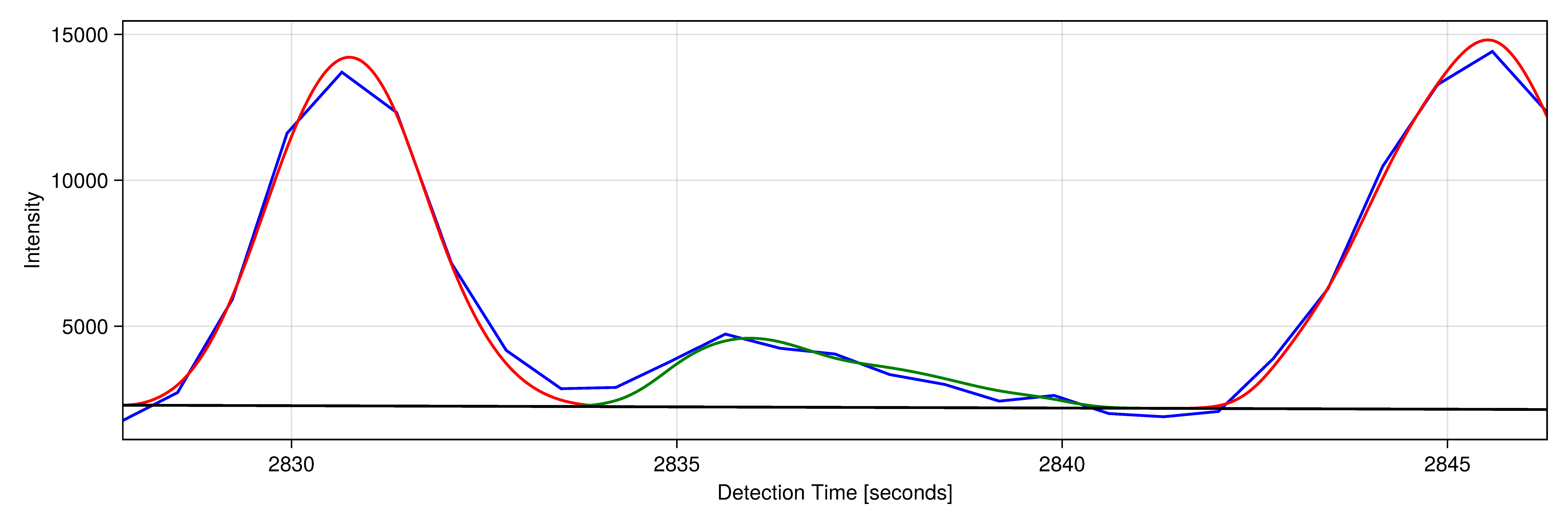 |
Biodiversity and Systematics
|
| Integrating genomic, chemical, and morphological data allows for unprecedented accuracy and precision in the delineation of species. We apply this approach to test the validity of controversial species and to screen for cryptic species. We also develop approaches for distinguishing between sibling species in the field that rely on non-invasively probing chemical compounds from the animals' integument. |
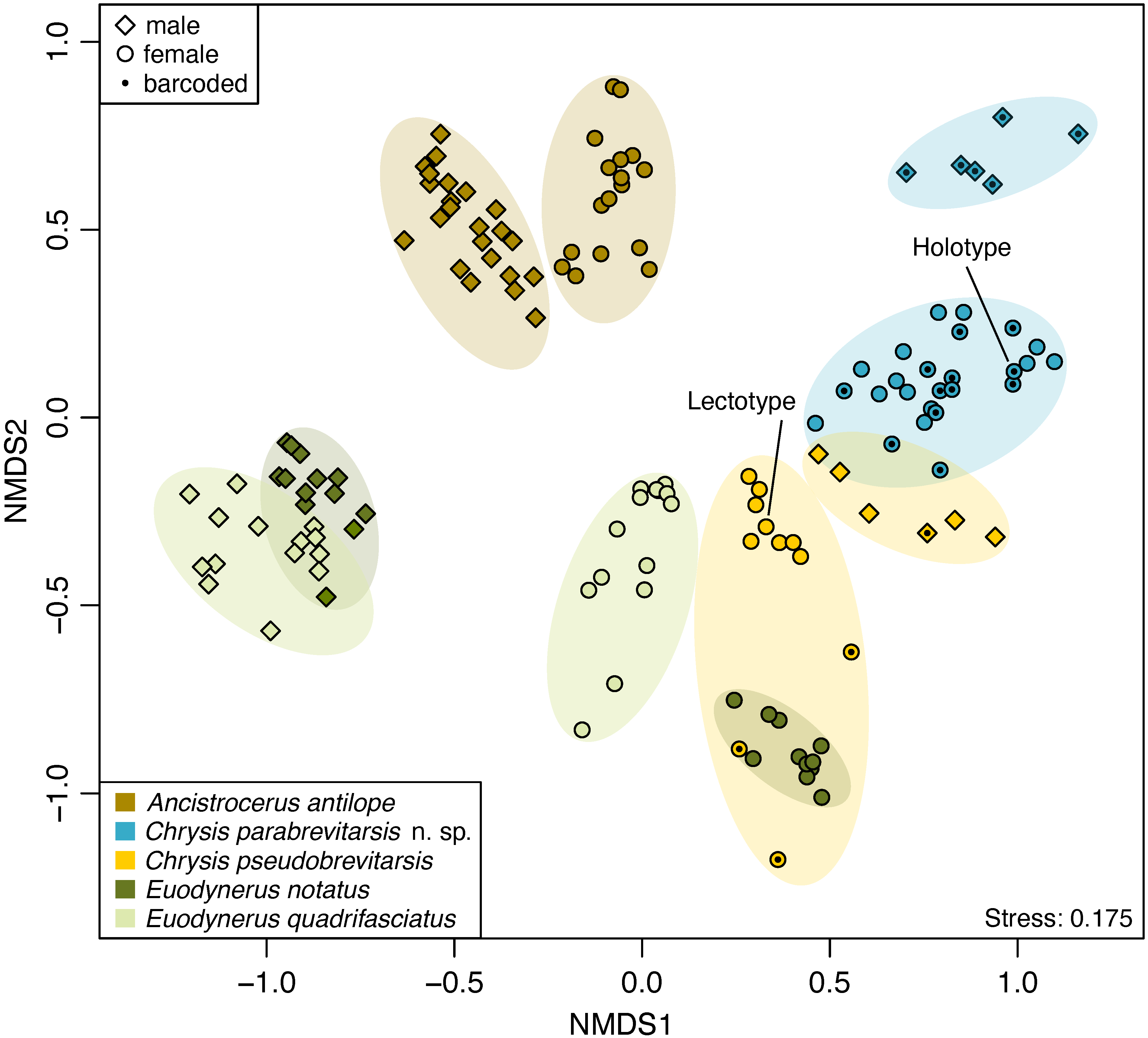 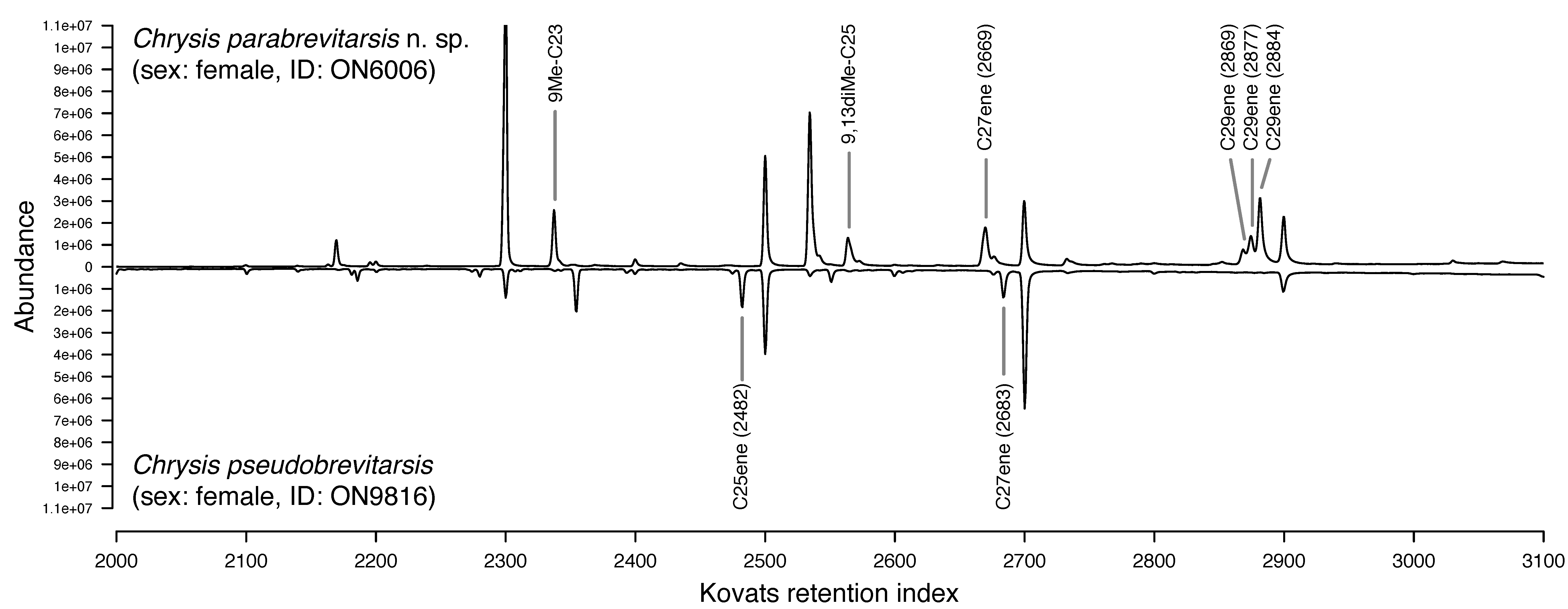 |
| |
|












